The theory behind the AMR DetecTool
Saving lives and costs with medication based on rapid AMR detection
1. Background of AMR
What is antibiotic resistance and how does it develop?
2. Mechanisms of bacterial resistance to antibiotics
A summary of the type of resistance and a detailed description of the mechanisms that the AMRDetecTool can detect
3. Spread of resistance
Spread of resistance from bacteria to bacteria & from patient to patient & and how to avoid or reduce it
4. The detection of resistance
The current possible methods and why LFIA is good
5. Resistance mechanisms the AMR DetecTool can detect
ESBLs: CTXM – MULTI, Carbapenemases: KPC, OXA, VIM, IMP, NDM, VAN A/B, OXA-Ab, 3GC
1. Background of AMR
Antibiotic resistance is the ability of bacteria to defeat the drugs designed to kill them. The increase and spreading of resistance can compromise the treatment of infectious diseases and other advanced therapeutic procedures as transplantation or anti-cancer therapy that involve immunosuppression and thus require robust anti-infective preventive therapies.
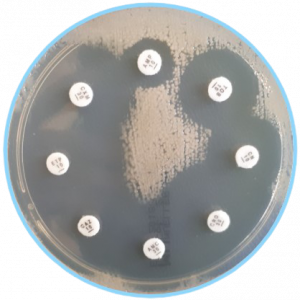
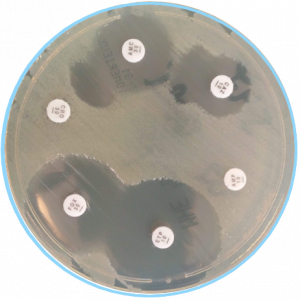
Bacteria change when exposed to antibiotics: using antibiotics may exert on bacteria a selective pressure leading to an increase in the prevalence of resistance.
The development of resistance is a natural phenomenon, but if antibiotics are not used properly, this process will speed up.
Origins and evolution of antibiotic resistance
Bacteria, other microbes, and plants produce small bioactive molecules, including antibiotics. Little is known about the natural biological functions of antibiotics, but they show extensive multifunctionality and are most likely involved in cell-cell signalling between bacteria and other organisms in the environment (fungi, plants, insects, livestock animals and even humans).
The discovery of antibiotics is one of the most significant health events of modern times, not just because of their ability to treat infectious diseases. In addition, studies with these compounds have often shown non-antibiotic effects, such as cardiovascular effects or immunosuppression.
The successful use of any therapeutic agent is compromised by the potential development of tolerance or resistance to that compound from the time it is first employed. A wide range of biochemical and physiological mechanisms may be responsible for resistance.
Resistance genes were not present before in the human bacterial pathogens, the source of the genes causing resistance to antimicrobials was the environmental microbiota, and the pathogenic bacteria acquired R-factors (resistance plasmids) from them.
Natural ecosystems, including not just soils but the human gut, contain a large number of elements that, upon transfer to a new host, can confer resistance to any type of antimicrobial. The gene that confers resistance in a human pathogen does not necessarily play the same role in its original host.
Those genes can be found nearly everywhere, including pristine ecosystems or wild animals not supposed to be in contact with antibiotics. This indicates that pollution with antibiotic resistance genes is widely spread and that resistance genes can persist even without positive selection pressure.
Bacteria typically coexist in complex multi-species communities, and the interactions within these communities can determine how bacteria respond to antibiotic exposure.
Bacteria secrete a vast variety of compounds ranging from signalling molecules involved in quorum sensing to exotoxins involved in virulence and competition. Secreted products have been shown to directly alter gene expression, metabolic processes and the growth dynamics of co-residing bacterial species, altering their ability to survive antibiotic exposure.
Interspecies interactions are especially common in polymicrobial infections where multiple species contribute to disease. These interactions not only affect the severity of infections but could also alter the exposure and physiological responses to antibiotics.
Clinical isolates from the same patient can protect each other from antibiotics, with the protective effects correlating with interspecies interactions that foster growth benefits.
Collective resistance, collective tolerance and exposure protection to antibiotics can also be provided through biofilm formation.
Sources:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3257838/
2. Mechanisms of bacterial resistance to antibiotic
- Change(s) in the antibiotic target sites – e.g. Vancomycin resistance
- Decrease of the membrane permeability – various antibiotics (e.g. aminoglycosides, fluoroquinolones, tetracycline)
- Efflux: the bacteria pump the antibiotics out – various antibiotics (beta-lactams, novobiocin,
erythromycin, fluoroquinolones, tetracycline, chloramphenicol, linezolid) - Antibiotic degradation: e.g beta-lactamase production
- Resistance genes are commonly found on plasmids or other mobile genetic elements (e.g. transposons)
- Plasmids can carry diverse resistance genes.
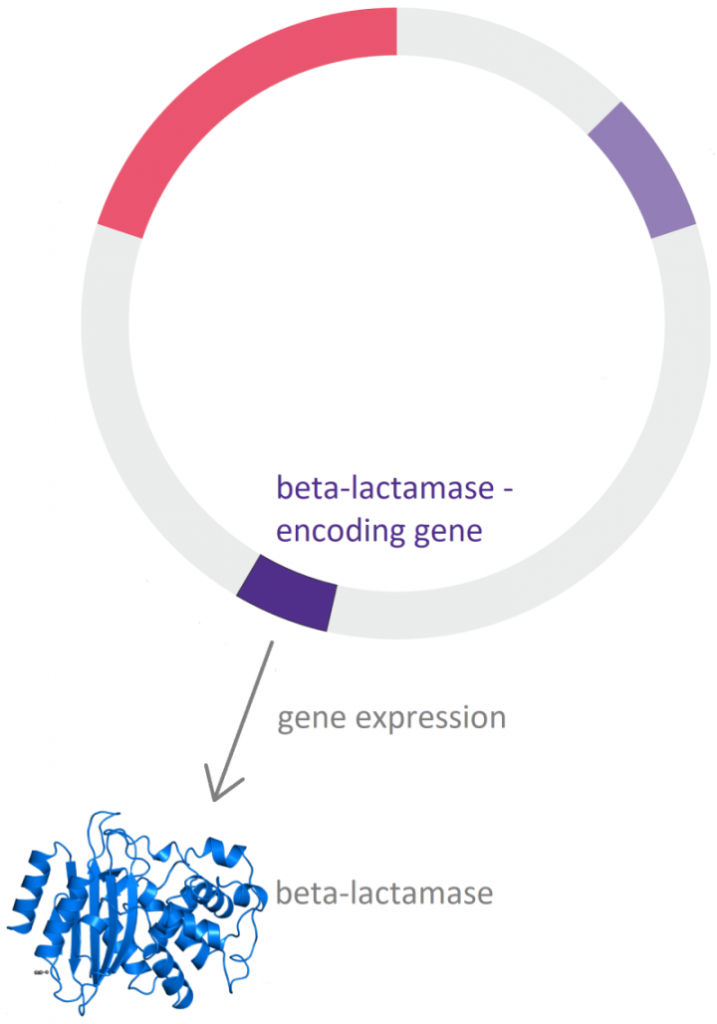
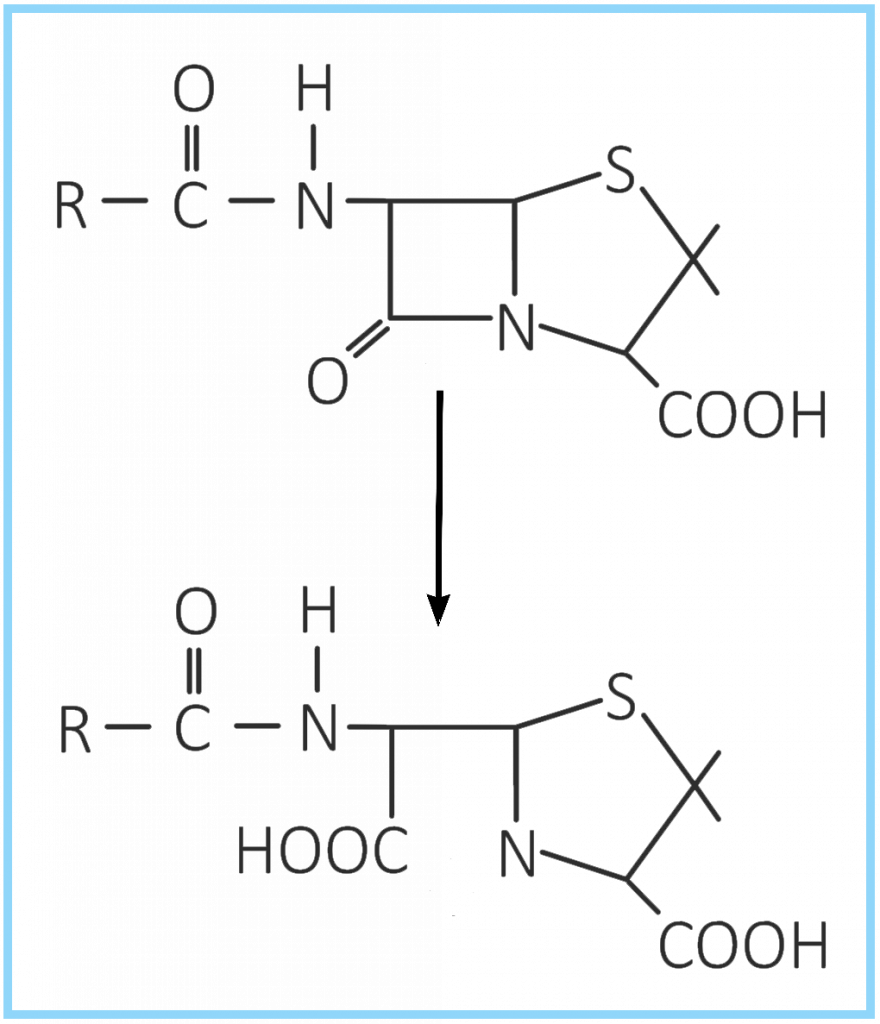
Beta-lactamases are enzymes produced by bacteria that inactivate the beta-lactam antibiotics by opening the ring at the amide bond.
Vancomycin resistance: change(s) in the antibiotic target sites
Vancomycin resistance is caused by an altered peptidoglycan terminus (d-ala-d-lac instead of the usual d-ala-d-ala), resulting in reduced vancomycin binding and failure to prevent cell wall synthesis.
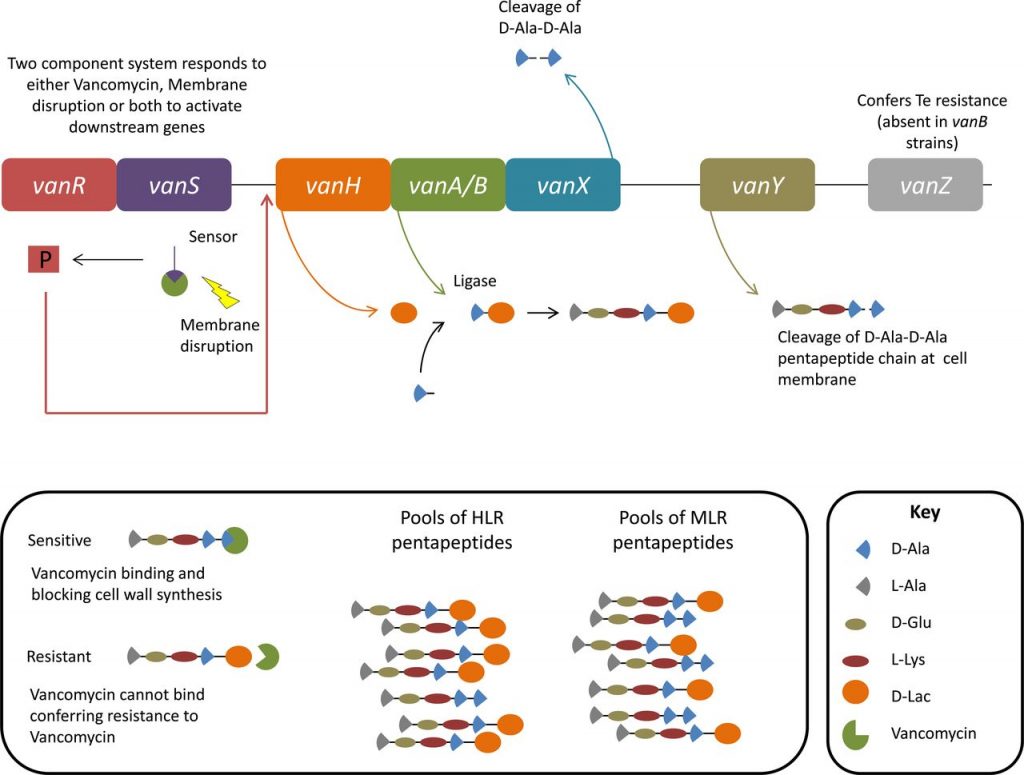
Sources:
3. Spread of resistance
From bacteria to bacteria:
Gene exchange is a universal property of bacteria.
The horizontal spread of genes is possible between the same and different species of Enterobacteriaceae.
During conjugation, the donor cell provides the plasmid to the recipient.
If the plasmid carries a resistant gene, the recipient cell becomes resistant as well.
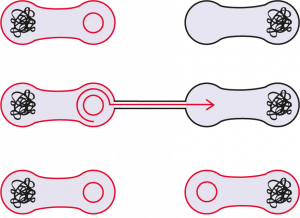
From patient to patient:
In case of inadequate hygiene, bacteria may spread from person to person
⇾ through contact with colonized people
- direct contact (touching)
- coughing and sneezing
- exposure to bodily fluids (such as close personal contact)
⇾ through contaminated devices or surfaces
Bacteria can live on surfaces for long periods of time. Bacteria can be picked up by contact with even everyday objects.
Bacteria may cause hospital-acquired infections: pneumonia, bloodstream infection, urogenital infection, skin and soft tissue infection, etc.
How to fight against the spreading of antimicrobial resistance
Combating antimicrobial resistance requires a multidisciplinary, collaborative, regulatory approach at international, national, community, hospital, individual and patient levels.
What to do at personal / community / national / international level:
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4149102/
What can you do in a hospital as a clinical / laboratory team member, and what to do a multidisciplinary team:
https://amrexpert.eu/fighting-amr-in-hospitals/
CDC recommends 12 steps to prevent antimicrobial resistance in a healthcare setting: https://www.cdc.gov/media/pressrel/r020326.htm
The WHO policy package to combat antimicrobial resistance:
https://www.who.int/bulletin/volumes/89/5/11-088435.pdf
4. The detection of resistance
Currently used methods for AMR detection
Polymerase chain reaction (PCR)
PCR enables rapid and exponential amplification of target DNA sequences using a forward and a reverse PCR primer and DNA polymerase in the presence of deoxyribonucleotides.
It can be used for detecting any genes that may be present within bacteria, as long as a DNA sequence is available for the whole or partial gene, which can be used to design the PCR primers.
Since the development of PCR, there have been several advances, which include real-time PCR (RT-PCR) and isothermal amplification, e.g., loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA).
Isothermal PCR techniques such as LAMP and RPA, differ from conventional or RT-PCR in that the whole process is performed at a constant temperature and does not require ramping up and down of temperatures.
Conventional PCR assays are well suited to detect the presence or absence of (resistance) genes but are less suited for the detection of point mutations within target genes unless subsequent Sanger sequencing is performed to detect these mutations.
Loop-mediated isothermal amplification (LAMP)
LAMP is a novel nucleotide amplification technique. The assay enables isothermal amplification of the target gene and thereby obviates the requirement for expensive thermal cycles. The method employs a single strand of DNA shaped like a dumbbell with loops at both ends. In the LAMP reaction gene amplification proceeds through the repetition of two types of elongation reactions that occur via loop regions. DNA having an inverse structure relative to the starting material is produced, and the starting material is formed again by the same reaction.
Matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS)
MALDI-TOF-MS can be used to identify specific bacteria types and resistance enzymes and has successfully been used to analyze a variety of bacterial strains. The complex instrumentation and time requirements may not offer a significant advantage relative to culture-based approaches.
___
Molecular AMR diagnostics detect resistance-coding genes or resistance-associated mutations in DNA extracted from purified bacterial isolates or directly from clinical samples. These methods can give much faster results than phenotypic methods, in some cases in a matter of minutes if a cultured bacterial colony is tested directly or within a few hours.
The disadvantages of some molecular methods are the price of reagents and instruments, and the technical expertise required.
AMR DetecTool
The AMR DetecTool is an affordable, sensitive, rapid and easy to use diagnostic system to detecting multidrug-resistant bacteria direct from clinical samples: blood culture, urine, rectal swab, bronchoalveolar lavage / tracheal aspirate.
The AMR DetecTool detects the enzymes directly from the patient’s sample, shortening the workflow for the detection of multidrug-resistant bacteria to 30 minutes.
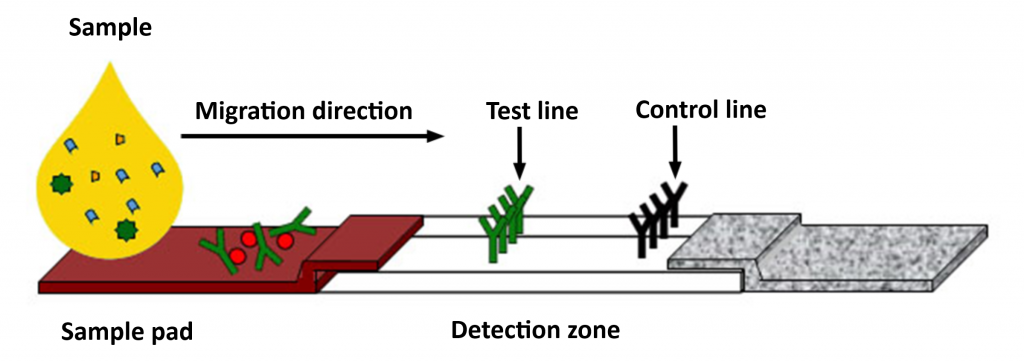
LFIA: Lateral Flow ImmunoAssay
The sample is applied at the end of the strip (sample pad), then migrates into the detection zone.
If the target is present in the sample, the antibodies will bind to it, recognising the test line and a control line, and coloured bands will appear.
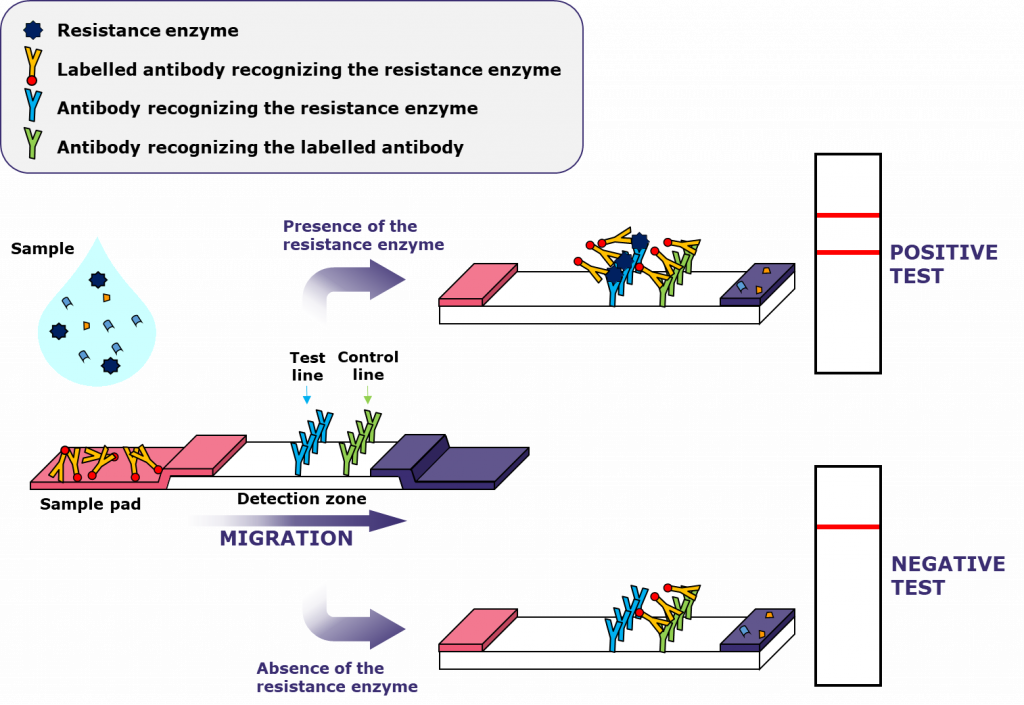
https://academic.oup.com/jac/article/73/4/909/4819243
https://journals.sagepub.com/doi/full/10.1177/2211068216680207
https://journals.asm.org/doi/10.1128/microbiolspec.ARBA-0011-2017
5. Resistance mechanisms the AMR DetecTool can detect
ESBL-s: CTX-M MULTI
ESBL (extended-spectrum beta-lactamase) production is becoming more common among Enterobacteriales.
The CTX-M-type extended-spectrum β-lactamase variants are the most frequently identified ESBLs.
ESBLs hydrolyze:
- penicillins
- 2nd, 3rd and 4th generation cephalosporins
- aztreonam
- (not cephamycins and carbapenems)
Carbapenemases: KPC, OXA, VIM, IMP, NDM
Carbapenemase production is more and more commonly reported in K. pneumoniae and has also been identified in other gram-negative pathogens including Pseudomonas aeruginosa.
Carbapenemases are beta-lactamases with versatile hydrolytic capacities.
Carbapenemases hydrolyze:
- penicillins
- cephalosporins
- monobactams
- carbapenems
Klebsiella with plasmid-mediated carbapenem resistance is a significant risk to hospitalized patients. The transfer of these resistance plasmids into E. coli is a significant public health threat because resistant E. coli may become part of the normal gut flora and thereby would be a source of infections in healthcare settings and the community. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3086234/)
VAN A/B: vancomycin-resistant Enterococci
Enterococci tend to persist in hospital environments, allowing for dissemination of resistance elements.
They are common colonizers.
Infections usually occur in immunosuppressed patients who have received multiple antibiotic treatments in the past.
Typically infection may be caused by enterococci:
- urinary tract infections
- intra-abdominal and pelvic infections
- skin and skin structure infections
- endocarditis
- central nervous system infections (rarely)
High risk of infection:
- chronic dialysis treatment
- severe underlying disease
- immunosuppressed condition
- use of invasive devices
- long hospital care
- previous or current antibiotic therapy
Van A is responsible for most of the human cases of VRE around the world and is mostly carried by E. faecium. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4521680/)
Enterococci carriers of VAN A gene are resistant to vancomycin and also teicoplanin, but the carriers of VAN B can be susceptible to teicoplanin.
OXA-Ab: OXA-type carbapenemases in Acinetobacter sp.
Acinetobacter species are significant pathogens in hospital-acquired and healthcare-associated infections worldwide.
Acinetobacter spp. may cause:
- ventilator-associated pneumonia
- bloodstream infection
- wound infection
- urinary tract infection
A. baumannii can develop resistance to multiple classes of antimicrobial agents.
The bacterium can be found anywhere in the hospital environment, can survive on dry surfaces for prolonged periods, often causes protracted outbreaks.
Risks of infection:
- stay in intensive care units
- burn injuries
- traumatology patients
- ventilated patients
- immunosuppressed condition
- serious underlying disease (chronic lung disease or diabetes mellitus)
3GC: third-generation cephalosporin resistance
3GC device for detection of determinants conferring third-generation cephalosporinase resistance (ESBLs (VEB, PER, GES, TEM, SHV), plasmid-encoded cephalosporinases (CMY, DHA)
Cephalosporins allow treating a variety of bacterial infections. Cephalosporin allergy is rare, and they generally cause few side effects. But the resistance against them is increasing continuously.
Sources:
https://pubmed.ncbi.nlm.nih.gov/12558458/
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3086234/
https://journals.asm.org/doi/10.1128/aac.01195-18?permanently=true
https://link.springer.com/article/10.1007/s40121-020-00350-1
Copyright 2020 – AMRDETECTOOL


